Solvation and Embedding Methods in Q-Chem
-
-
SM8, SM12, SMD, COSMO, C-PCM, SS(V)PE, IEF-PCM, CMIRS, and more
-
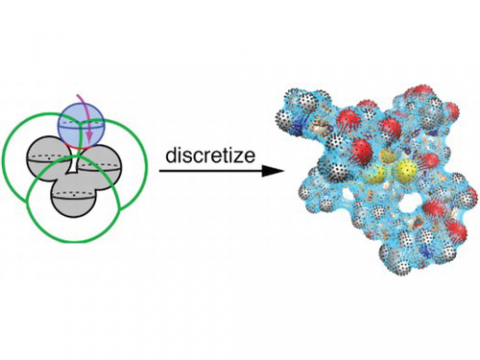
Intrinsically smooth discretization of the solute/continuum interface
-
Poisson equation solver for anisotropic dielectric boundary conditions
-
-
Effective fragment potential (EFP) for modeling explicit solvent with polarizable embedding
-
Built-in library of effective fragments and user-defined potentials
-
Available for DFT and wave function-based ground and excited state methods
-
Fragmented EFP scheme for macromolecules
-
-
Stand-alone QM/MM capabilities
-
Available for ground- or excited-state QM treatments
-
“Yin-Yang atom” approach to defining the QM/MM boundary
-
Integration with PCM models (QC/MM/PCM)
-
Many-body expansion for incorporating solvent at a QM level
-
-
Interface to CHARMM and GROMACS
-
-
Projected-based density embedding for DFT-in-DFT or WFT-in-DFT
-
Frozen-Density embedding theory (FDET) and polarizable embedding (PE via EFP) for Algebraic Diagrammatic Construction (ADC) scheme
-
Want to try Q-Chem?